New research reveals the architecture of an essential mRNA processing machinery.
DNA contains information needed to make proteins – the basic building block of a cell. The information in the DNA is first transcribed into an intermediate messenger RNA (mRNA), which is then used as the template to make proteins. In a nutshell, this is the central dogma of molecular biology.
Our cells consist of numerous molecular machines that are made up of proteins and nucleic acids. These large molecular machines carry out most of the tasks necessary for cell survival including transcription of DNA into mRNA, additional processing of the mRNA and translating the information in the mRNA into proteins. An essential and poorly understood step in the central dogma involves the cleavage of an mRNA and the addition of a polyadenosine or poly(A) tail. This is carried out by a complex multi-protein machinery known as the Cleavage and Polyadenylation Factor (CPF).
The mRNA poly(A) tail is essential for the efficient export of the mRNA into the cytoplasm and subsequently has an influence on mRNA translation into proteins. Not surprisingly, defects in mRNA poly(A) tail addition are associated with several diseases, including cancer. Despite the central role of poly(A) tails in the life of an mRNA, the mechanistic underpinning for the poly(A) addition by CPF has remained elusive.
Gates Cambridge scholar Anantha Kumar [2015] and EMBO fellow Dr Ana Casañal worked with a team of scientists led by Dr Lori Passmore in the MRC Laboratory of Molecular Biology in Cambridge to study this. They now report a major advancement in our understanding of the structure and function of CPF and how poly(A) tails are added. The research, Architecture of eukaryotic mRNA 3-end processing machinery was published in the journal Science this week.
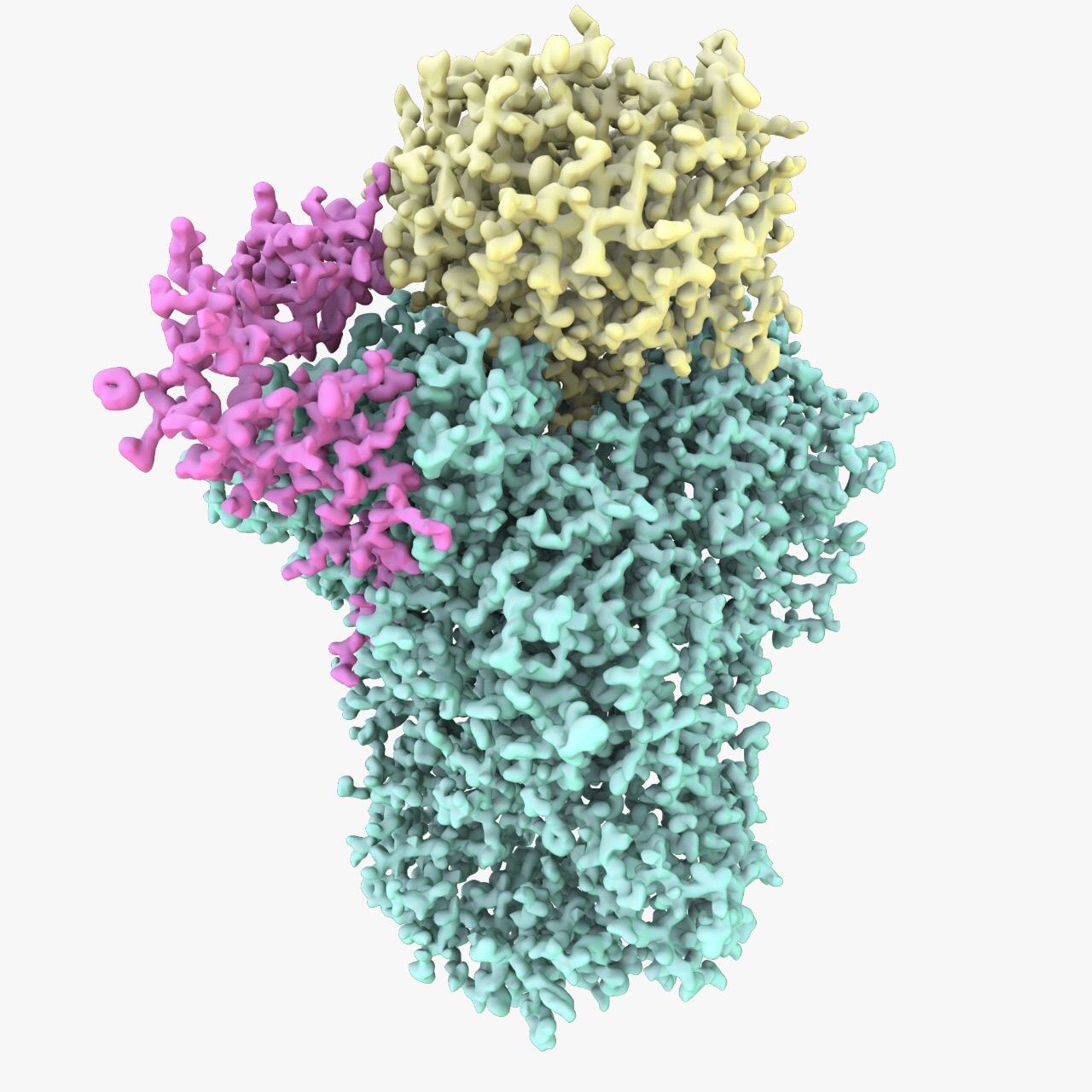
Experiments, done in collaboration with Dr Carol Robinson’s group at the University of Oxford, revealed that CPF is assembled from 14 different protein subunits organised into three modules. These modules are segregated around the cleavage, polymerase and phosphatase enzymes of CPF. “Although we have known the identities of the proteins that make up CPF for many years, it was not clear how they assemble into one complex. Our interaction map provides a new conceptual framework to understand how the CPF functions. It also allowed us to produce individual modules for structural studies,” explains Dr Passmore.
The polymerase module identified by the team consists of five protein subunits, including the polymerase enzyme Pap1, and is also present in humans. Anantha [2015], who is a third year PhD student in Biological Science at Churchill College, said: "Pap1 can add a poly(A) tail to a substrate mRNA by itself. But in the cell, Pap1 exists as a part of a bigger complex along with several other protein subunits. We were really tantalised by the roles of these other subunits in poly(A) tail addition."
"We took a biochemical and structural biology approach in order to clarify the interactions, architecture and functions of the subunits," added Dr Casañal.
The team purified the polymerase module and captured it in vitreous ice in its native state. They then imaged it using a transmission electron microscope – this technique, known as cryo-EM, was awarded the Nobel Prize in Chemistry this year. This led to the determination of an atomic-resolution structure of the complex. For the first time, the team was able to visualise the intricate and extensive interactions between the different protein subunits that hold the complex together. To their surprise, the structure of the polymerase module resembles other protein complexes in the cell that are involved in RNA splicing and DNA repair. Furthermore, by combining the structure of the polymerase module with previous data from other groups, the team provides a new explanation for how the influenza A virus could take control of the host cell. “We were trying to understand how this fundamental molecular machine works in the cell. We were delighted to also gain insight into influenza infection,” explained Anantha.
By combining data from biochemical experiments, cryo-EM and mass spectrometry, the team found that the polymerase module acts a hub for facilitating specific and efficient poly(A) tail addition. One can think of the polymerase module as similar to the international space station docking and berthing. It's a busy hub where all the interactions take place," added Anantha and Dr Casañal who are currently focussing on trying to get detailed structural information about such interactions.
“This work is a big achievement. It opens the door to future studies directed at understanding how expression of genes is controlled by this cellular machine,” Dr Passmore concluded.
The research was funded by the Gates Cambridge, MRC, the European Research Council (ERC), and EMBO. The researchers used electron microscope facilities at the MRC LMB and the Diamond Light Source.
*Picture credit: Ana Casañal and Thomas Martin.

Ananthanarayanan Kumar
- Alumni
- India
- 2015 PhD Biological Science @ MRC LMB
- Churchill College
Structural studies of molecular machines and their assembly are my primary research interest. Ever since my Bachelors thesis project at Trans-Membrane Trafficking lab at OIST (Japan), I have been amazed by the tricky membrane proteins and the complexity behind their structural studies. My Masters thesis work at the Bio-membrane Functions Lab in Nagoya University (Japan) involved the functional and structural analysis of a flagellar inner membrane protein from Vibrio alginolyticus. During my Masters, I worked closely with Imada sensei's lab at Osaka University to learn X-ray crystallography. Next, as a summer intern at the Mancini lab at STRUBI (Oxford University), I developed a desire to know more about structural dissection of pathways associated with gene regulation such as Chromatin remodelling and mRNA processing. This led me to pursue my PhD research at the Passmore lab in MRC LMB with a GATES Cambridge scholarship. During my PhD, using an integrative structural biology approach, I unraveled the structural architecture of the eukaryotic mRNA 3' end polyadenylation machinery (https://doi.org/10.17863/CAM.43327). I specialise in using a combination of in vitro biochemistry, cryo-EM and NMR to study the structure and dynamics of protein-RNA complexes associated with fundamental cellular processes. I am now pursuing my postdoc research at the Pyle lab, MCDB, Yale University.
Previous Education
SASTRA University
Nagoya University












